Scaffan
Main goal of application Scaffan is quantitative description of scaffold based on image slice.
The algorithm can be separated into two steps. First step is lobulus segmentation. In this step the region of segmentation. Based on user interactivity the rough localization of examined lobulus is known. This information is used as initial step for iterative segmentation algortihm based on Active Contours without Edges described in Snakes and Neila. Output of this process is delineation lobulus boundary and central vein.
Second step is lobulus area description. Input of this process is the lobulus area. Texture in this area is described with several methods. The Gray Level Cooccurence Matrix texture features described in Haralick were used as main texture features. Based on this texture features the Sinusoid Integrity can be estimated.
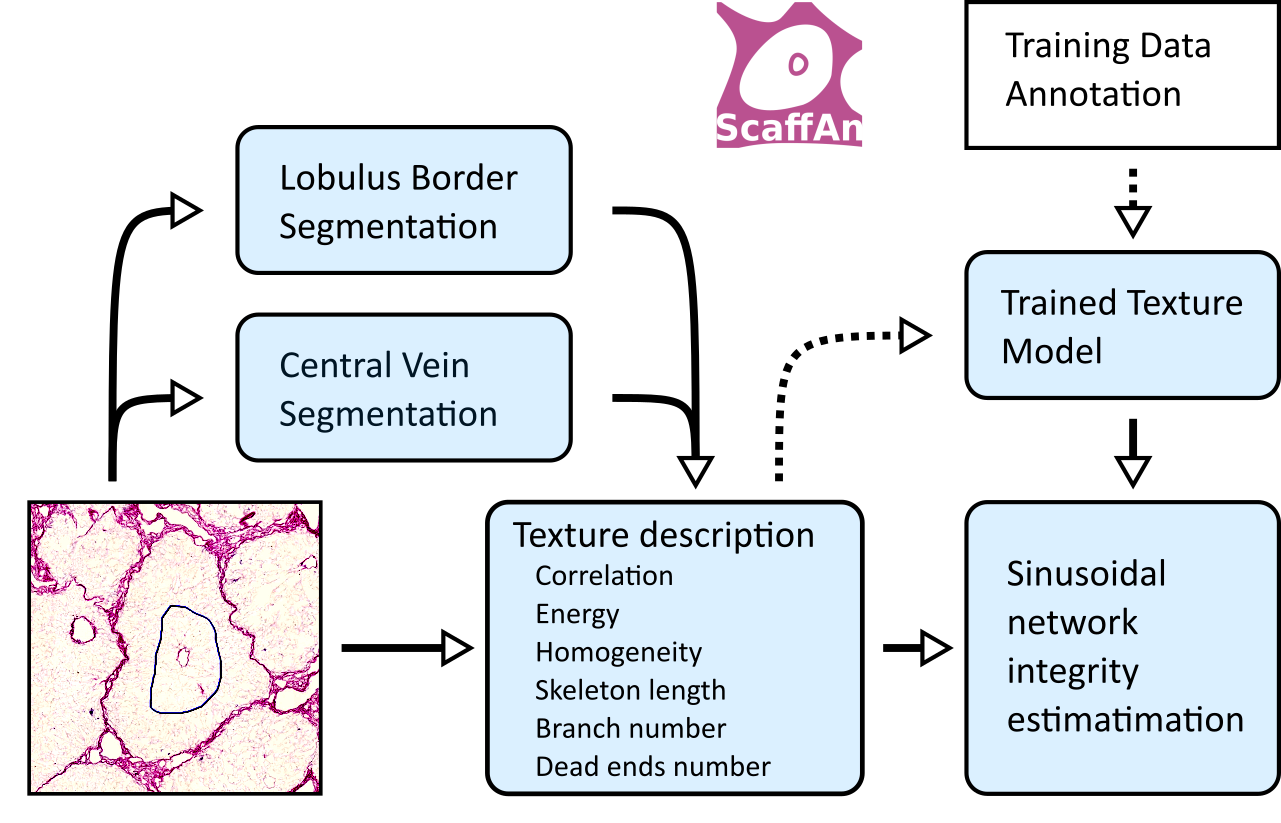
Application
Application Scaffan is open-source software hosted on GitHub and written in (Python)[#python]. The packages Numpy, Scipy, Scikit-image are used for general image processing. The morphsnakes package (Neila and Avarez) are used for segmentation tasks. For machine learning the Scikit-learn is used.
Input data
Input data can be stored in Hamamatsu .ndpi file format. The OpenSlide library (described in
Analyzing huge pathology images with open source software
and
Open source tools for digital microscopy data
)
is used to read the data. Image data format is described in
OpenSlide Hamamatsu image format.
Lobulus segmentation
Initial step of lobulus segmentation is user interactivity over the image slice. User is expected to draw closed curve around the central vein of examined lobulus. This curve is used in several moments of processing.
At first, the input image is cropped around input curve with margin size defined by Annotation Margin parameter (default vaule is 180%). The image area should be big enough to cover whole area of lobulus. The resolution of image is changed to 1.82um. It is enough to keep all important structures visible and also keep the computation time low.
The preprocessing step of lobulus segmentation is to use Hessian based Frangi filter Frangi, Kroon to supress the high-frequency signal in lobulus area and to make the lobulus border more dominant. Then a method Morphological Active Contours without Edges described in Casselles, Neila and Avarez is used to extract outer boundary of liver lobulus.
Due to lower contrast the segmentation of Central Vein is more complex task. We use the texture analysis to detect roughly the area of Central Vein. Gradient magnitude filtration using Gaussian derivatives combined with the Frangi filter is used to have more details of the shape of Central Vein.
Lobulus analysis
To describe texture properties in the Gray Level Cooccurence Matrix texture features described in Haralick and Texture Analysis Additionally the manually crafted features were used.
Segmentation evaluation
To evaluate lobulus area segmentation the manual annotation of data was performed. Our semi-automatic segmentation method was compared with manual segmentation of lobulus border and central vein. The Jaccard simmilarity coefficient (also known as Intersection over Union) ( Jaccard and Levandowsky ) was used to measure similarity between samples.
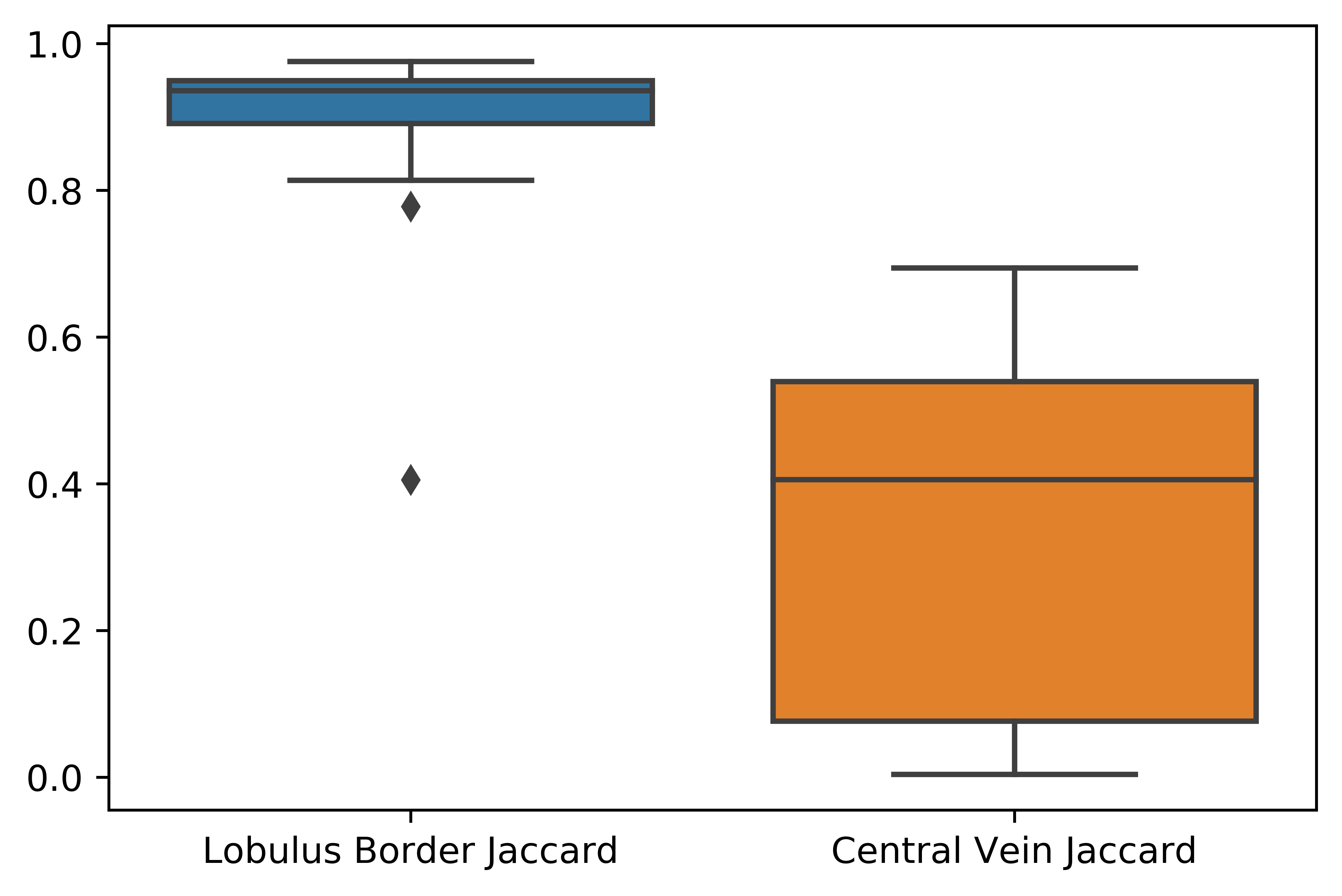
Lobulus border and central vein segmentation evaluation by Jaccard Index. Whiskers proportionaly extends low and high quartiles by 1.5 of the interquartile range (IQR). Points outside this range are be identified as outliers.
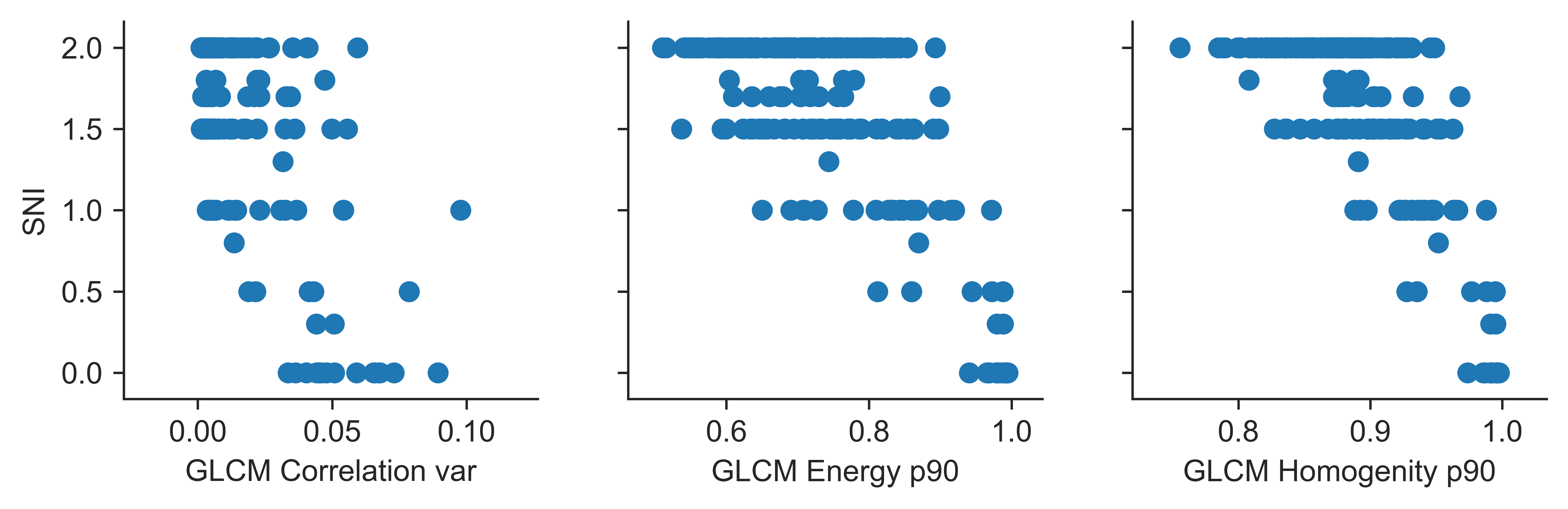
Sinusoid ingegrity evaluation
Reference-style:
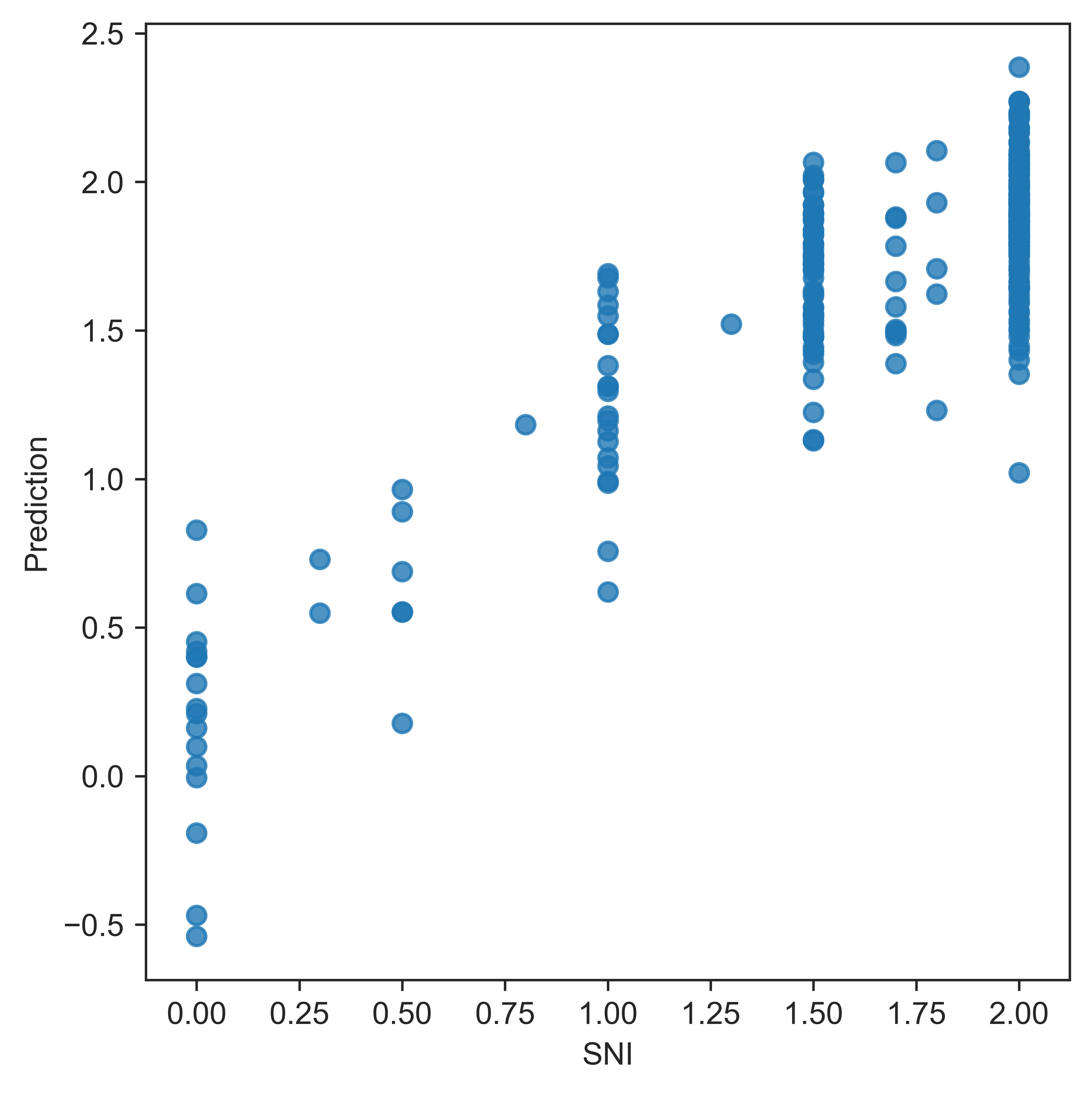
b
Annotation
Geometric annotation is done with Freehand Region tool. We use fallowing color code:
- blue - PV selection
- green - PV selection with additional black annotation of portal vein and lobulus
- black - Additional annotation of portal vein and lobulus border
- magenta - training data for empty space
- red - training area for septum
Written in Annotation Details field
SNI: Sinusoidal Integrity <0,2>SQZ: Squeeze 0-no squeeze, 1-squeezed (maybe different types? anisotropic(getting suqeezed), isotropic (getting small))CMP: Compactness <0,1>IFE: Image Focus Error <0,1>AIS: Anisotropy <0,1> 0 for random orientation, 1 for big directionalityCVM: Central Vein Missing, 0 Central Vein is in the image, 1 - Central Vein is not in the imageICE: Image Contrast Error <0,1>, 0 - image contrast is ok, 1 - image contrast is lowSCE: Slice Contamination Error <0,1>
texture problem - local / global
User interface
Scaffan work with image data in .ndpi format and .ndpa annotation . This annotation can be prepared in Hamamatsu
NDP.view 2 application. Scaffan require to annotate each lobulus by closed curve around central vein.
References:
Frangi
A. Frangi, W. Niessen, K. Vincken, and M. Viergever. “Multiscale vessel enhancement filtering,” In LNCS, vol. 1496, pages 130-137, Germany, 1998. Springer-Verlag
Kroon
Kroon, D.J.: Hessian based Frangi vesselness filter.
Neila
A Morphological Approach to Curvature-based Evolution of Curves and Surfaces, Pablo Márquez-Neila, Luis Baumela and Luis Álvarez. In IEEE Transactions on Pattern Analysis and Machine Intelligence (PAMI), 2014, DOI 10.1109/TPAMI.2013.106
Alvarez
Morphological Snakes. Luis Álvarez, Luis Baumela, Pablo Márquez-Neila. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2010 (CVPR10).
Casselles
Geodesic Active Contours, Vicent Caselles, Ron Kimmel and Guillermo Sapiro. In International Journal of Computer Vision (IJCV), 1997, DOI:10.1023/A:1007979827043
Chan
Active Contours without Edges, Tony Chan and Luminita Vese. In IEEE Transactions on Image Processing, 2001, DOI:10.1109/83.902291
Python
@book{van1995python,
title={Python tutorial},
author={Van Rossum, Guido and Drake Jr, Fred L},
year={1995},
publisher={Centrum voor Wiskunde en Informatica Amsterdam, The Netherlands}
}
Numpy
@book{book,
author = {Oliphant, Travis},
year = {2006},
month = {01},
pages = {},
title = {Guide to NumPy}
}
Scipy
@Misc{scipy,
author = {Eric Jones and Travis Oliphant and Pearu Peterson and others},
title = {SciPy: Open source scientific tools for Python},
year = {2001--},
url = "http://www.scipy.org/",
note = {[Online; accessed ]}
}
Scikimage
@article{van2014scikit,
title={scikit-image: image processing in Python},
author={Van der Walt, Stefan and Sch{\"o}nberger, Johannes L and Nunez-Iglesias, Juan and Boulogne, Fran{\c{c}}ois and Warner, Joshua D and Yager, Neil and Gouillart, Emmanuelle and Yu, Tony},
journal={PeerJ},
volume={2},
pages={e453},
year={2014},
publisher={PeerJ Inc.}
}
Sciklearn
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V.
and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P.
and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and
Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
Sklearn API
@inproceedings{sklearn_api,
author = {Lars Buitinck and Gilles Louppe and Mathieu Blondel and
Fabian Pedregosa and Andreas Mueller and Olivier Grisel and
Vlad Niculae and Peter Prettenhofer and Alexandre Gramfort
and Jaques Grobler and Robert Layton and Jake VanderPlas and
Arnaud Joly and Brian Holt and Ga{\"{e}}l Varoquaux},
title = {API design for machine learning software: experiences from the scikit-learn
project},
booktitle = {ECML PKDD Workshop: Languages for Data Mining and Machine Learning},
year = {2013},
pages = {108--122},
}
Jaccard
https://nph.onlinelibrary.wiley.com/doi/abs/10.1111/j.1469-8137.1912.tb05611.x
Levandowsky
Levandowsky, Michael; Winter, David (1971), “Distance between sets”, Nature, 234 (5): 34–35
https://www.nature.com/articles/234034a0
Texture Analysis
Tuceryan, M., & Jain, A. K. (1998). Texture Analysis. In The Handbook of Pattern Recognition and Computer Vision (2nd Edition). Retrieved from http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.38.5980
Haralick
Haralick, R.M., “Statistical and Structural Approaches to Texture,” Proceedings ofthe IEEE, 67, pp. 786-804, 1979.
Snakes
Snakes: Active contour models (1988) by Michael Kass , Andrew Witkin , Demetri Terzopoulos Venue: INTERNATIONAL JOURNAL OF COMPUTER VISION Citations: 3874 - 17 self
https://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.124.5318
OpenSlide Hamamatsu image format
https://openslide.org/formats/hamamatsu/
Open source tools for digital microscopy data
@article{Khushi2013,
abstract = {Background: Virtual microscopy includes digitisation of histology slides and the use of computer technologies for complex investigation of diseases such as cancer. However, automated image analysis, or website publishing of such digital images, is hampered by their large file sizes.Results: We have developed two Java based open source tools: Snapshot Creator and NDPI-Splitter. Snapshot Creator converts a portion of a large digital slide into a desired quality JPEG image. The image is linked to the patient's clinical and treatment information in a customised open source cancer data management software (Caisis) in use at the Australian Breast Cancer Tissue Bank (ABCTB) and then published on the ABCTB website (http://www.abctb.org.au) using Deep Zoom open source technology. Using the ABCTB online search engine, digital images can be searched by defining various criteria such as cancer type, or biomarkers expressed. NDPI-Splitter splits a large image file into smaller sections of TIFF images so that they can be easily analysed by image analysis software such as Metamorph or Matlab. NDPI-Splitter also has the capacity to filter out empty images.Conclusions: Snapshot Creator and NDPI-Splitter are novel open source Java tools. They convert digital slides into files of smaller size for further processing. In conjunction with other open source tools such as Deep Zoom and Caisis, this suite of tools is used for the management and archiving of digital microscopy images, enabling digitised images to be explored and zoomed online. Our online image repository also has the capacity to be used as a teaching resource. These tools also enable large files to be sectioned for image analysis.Virtual Slides: The virtual slide(s) for this article can be found here: http://www.diagnosticpathology.diagnomx.eu/vs/5330903258483934. {\textcopyright} 2013 Khushi et al.; licensee BioMed Central Ltd.},
author = {Khushi, Matloob and Edwards, Georgina and de Marcos, Diego Alonso and Carpenter, Jane E. and Graham, J. Dinny and Clarke, Christine L.},
doi = {10.1186/1746-1596-8-22},
issn = {17461596},
journal = {Diagnostic Pathology},
keywords = {Caisis,Deep Zoom,Digital slides,NDPI-Splitter,Snapshot Creator,Virtual microscopy},
number = {1},
pages = {1--7},
title = ,
volume = {8},
year = {2013}
}
Analyzing huge pathology images with open source software
@article{Deroulers2013,
abstract = {Background: Digital pathology images are increasingly used both for diagnosis and research, because slide scanners are nowadays broadly available and because the quantitative study of these images yields new insights in systems biology. However, such virtual slides build up a technical challenge since the images occupy often several gigabytes and cannot be fully opened in a computer's memory. Moreover, there is no standard format. Therefore, most common open source tools such as ImageJ fail at treating them, and the others require expensive hardware while still being prohibitively slow.Results: We have developed several cross-platform open source software tools to overcome these limitations. The NDPITools provide a way to transform microscopy images initially in the loosely supported NDPI format into one or several standard TIFF files, and to create mosaics (division of huge images into small ones, with or without overlap) in various TIFF and JPEG formats. They can be driven through ImageJ plugins. The LargeTIFFTools achieve similar functionality for huge TIFF images which do not fit into RAM. We test the performance of these tools on several digital slides and compare them, when applicable, to standard software. A statistical study of the cells in a tissue sample from an oligodendroglioma was performed on an average laptop computer to demonstrate the efficiency of the tools.Conclusions: Our open source software enables dealing with huge images with standard software on average computers. They are cross-platform, independent of proprietary libraries and very modular, allowing them to be used in other open source projects. They have excellent performance in terms of execution speed and RAM requirements. They open promising perspectives both to the clinician who wants to study a single slide and to the research team or data centre who do image analysis of many slides on a computer cluster.Virtual slides: The virtual slide(s) for this article can be found here:. http://www.diagnosticpathology.diagnomx.eu/vs/5955513929846272. {\textcopyright} 2013 Deroulers et al.; licensee BioMed Central Ltd.},
archivePrefix = {arXiv},
arxivId = {1709.02293},
author = {Deroulers, Christophe and Ameisen, David and Badoual, Mathilde and Gerin, Chlo{\'{e}} and Granier, Alexandre and Lartaud, Marc},
doi = {10.1186/1746-1596-8-92},
eprint = {1709.02293},
issn = {17461596},
journal = {Diagnostic Pathology},
keywords = {Digital pathology,Image processing,ImageJ,NDPI,Systems biology,Virtual slides},
number = {1},
pages = {1--8},
title = ,
volume = {8},
year = {2013}
}